| Service | University of California | Non-Profit | For-Profit |
|---|---|---|---|
| Protein ID LC-MS/MS Q-Exactive | $169.00 | $259.00 | $308.00 |
| Extra Data Analysis | $70.00 | $108.00 | $128 |
* These are bundled prices, for our official prices please see http://proteomics.ucdavis.edu/prices-jan-2015
This Service includes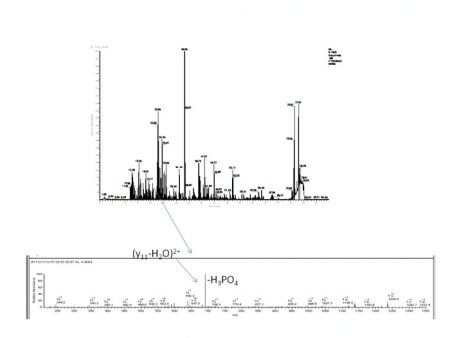
- A quality control run at 125 fmol
- Proteolytic digestion (Trypsin, other proteases avaliable)
- Database search
- PTM search via x!tandem, SEQUEST or Mascot, Localization scores for Phosphorylations
- Manual Verification
We identify proteins PTM’s by digesting the proteins into peptides, analyzing the peptides using tandem mass spectrometry (LC-MS/MS), and then searching for modifications using X!tandem (our default search engine), SEQUEST, or specialized PTM software (see above). We will verify the modification by hand and give you our opinion of their likelihood of being correct along with the scores from the specific programs we use.
Some Modifications are easier to find than others. We can look for specific modifications (Phosphorylations, etc..) or scan your data for unknown modifications, e.g. modifications that you did not know about (pretty cool no?). Usually modifications we can look for can be found in the unimod database
Your chances of detecting the modification you are interested in are completely dependent on the stability of that modification, how that modification affects peptide fragmentation in a mass spectrometer, the stoichiometry of the modification and numerous other variables. Basically, we don’t have a clue most of the time whether we will be successful. You have to try it and see. General rules to live by are, more material the better and expect to try it a few times.
Analyzing some specific modifications require specialized expertise and equipment that we do not have (such as carbohydrates). We can provide you with some names if you need that type of analysis.
Papers
Phosphorylation



